Terminology Sharing White paper
Proposed Profile: Terminology Sharing White paper
- Proposal Editor: Christel Daniel (AP-HP, INSERM, Paris)
- Editor: Christel Daniel (AP-HP, INSERM, Paris), Ana Esterlich (GIP-DMP), François Gareil (Thales), Charles Rica (GIP DMP), Karima Bourquard (GMSIH), Jean Delahousse (Mondeca), Pierre Zweigenbaum (LIMSI, CNRS)
- Date: N/A (Wiki keeps history)
- Version: N/A (Wiki keeps history)
- Domain: <IT infrastructure>
Summary and Scope
To achieve interoperability within and across disparate healthcare IT systems, terminological resources for indexing both narrative and structured clinical information need to be synchronized across the various applications (at a local, or regional, or national, or international level).
Existing standards (e.g Common Terminology Services) address the issue of sharing terminology resources.
This white paper extends the Integration Profile “Sharing Value Sets" to address the following issues:
- Updating Value Sets ("how mapping between terminological resources (Code Systems) and data model (e.g. CDA r2-based templates or DICOM objects) are defined and revised")
- Distributing Terminology mappings (how mapping between existing terminological resources (Code Systems)are defined, distributed and revised)
- Terminology correction or authoring("how Terminology (Code Systems) users submit unambiguous requests for corrections and extensions to Terminology providers (e.g SNOMED CT IHT SDO)and how revisions to content are identified, distributed and integrated into running systems").
At last, the “Terminology Sharing” profile will define business scenarios for Terminology Services for Narrative documents indexing and mining(how terminological resources (Code Systems)are distributed to IT applications for efficient use of Natural Processing Language (NLP) algorithms.
This requires that:
- 1) Terminology Providers (using Terminology Creator Sources) distribute terminological resources (Code Systems) to Terminology Administrators in charge of terminology servers(Terminology Repositories/Registries). Then, Terminology Authors/curators are defining and maintaining within these terminology servers (Terminology Source/Mapping/Value Sets Repositories/Registries)the relationships between Code Systems and domain content models (Value Sets authoring) and the relationships between different terminological resources (Code Systems)(Terminologies mappings)
- 2)Terminology Administrators of terminology servers(Terminology/Mapping/Value Sets Repositories/Registries) distribute updated inter-related Code Systems and Value Sets to Terminology Users using IT applications (Terminology Consumers).
Task force:
- HSSP and CTS2
- In France many hospitals are deploying new generation of Clinical Information Systems and the government conducts a project of a national scale a PHR (Personal Health Record). The GIP-DMP (french public organization of the PHR), the GMSIH (The Association in Charge of the Modernization of the Healthcare Information Systems), researchers from the INSERM (The National Institute of Health and Medical Research), CNRS (National Center for Scientific Research), and the Association of Hospitals of Paris containing more then 40 hospitals are willing to put efforts into this profile. There is a strong interest from the industry side, namely companies such as Thales, Mondeca, and DBmotion.
The main challenges are proposing an operational solution to share the semantic of EHR (narrative documents, structured documents (based on domain content models such as CDA r2 or DICOM objects, etc) avoiding inconsistent implementations and heterogeneity of technical solutions across domains.
The Problem
1) Semantic interoperability and semantic mining.
Clinical information systems (CIS) usually offer solution for basic workflow needs and clinical documentation. Existing de facto CIS contain heterogeneous data, much of them still in an unstructured form, such as narrative case notes and correspondence. There is often a lack a complete view of the clinical data since these data are scattered between different sub-systems within different departments or healthcare provider organizations and both narrative and structured data are not indexed thanks the consistent use of a terminology system. Systems that have to share clinical data and information have to share also well defined and unambiguous knowledge of the meaning of these data. Accessing and utilizing terminology resources are a common and necessary function for many healthcare IT applications. New generation of CIS take benefits of health informatics knowledge-representation contributions such as biomedical ontologies (e.g OBO fundry, SNOMED-CT) and data model for structured medical information (such as HL7 Clinical Document Architecture Release 2 or archetypes) Significant advance in EHR interoperability is maturing at the level of specifications but are still to demonstrate wide-scale adoption and success at providing a seamless clinical information environment that can support shared care at a national or regional level [1,2]. The coupling of terminology systems and information models does not only support the goal of semantic interoperability, but, when using formal ontologies, it also makes clinical information subject to formal, classification-based reasoning and allows the use of novel data mining algorithms. A coherent knowledge view of distributed and heterogeneous clinical data is also useful for their connection with medical knowledge (e.g. published medical literature and clinical guidelines). There is a challenge to define the best combination of EHR information models, domain content models (archetypes, templates based on CDA r2, DICOM objects, etc) and ontologies to harmonise the clinical data from multiple sites and systems and link these data to relevant medical knowledge.
2) Binding clinical data and terminology resources
a) Terminology Systems Issues concerning the representation of knowledge and the provision of terminology services in clinical and research contexts have been intensively discussed in the Medical Informatics community over the last two decades [3-7]. Existing bio-medical terminology resources (different systems of classifications, terminologies and ontologies) have been created in purely application- and purpose-driven contexts. Addressing semantic interoperability is increasingly being fulfilled by the Open Biomedical Ontologies (OBO) foundry approach for the basic biomedical sciences [8] and by the maturation of the clinical SNOMED CT terminology [9-11]. Formal biomedical ontologies define classes or types of entities, whose properties are expressed by a formal semantics using formalisms such as Description Logics [12]. The entities represented in formal ontologies are abstract types, not language terms. Although, individuals (e.g. the pneumonia of patient X, the fever of patient Y) are not represented in the ontology, the main purpose of ontology is to classify individual entities by defining and organizing the semantic types they are instances of [13,14]. Taxonomic reasoning is the most important mechanism of machine inference applied to formal ontologies.
b) Indexing structured data : binding domain content model to terminology system The relation between clinical information models and ontologies has recently received an increased attention in the context of HL-7 Version 3, clinical archetypes, and SNOMED CT[15]. This relation has been problemized e.g. by experiences from large scale implementation attempts such as the UK Connecting for Health project[16]. A consensus regarding the framework needed to deal with this problem has been reached as one of the outcomes of the EU founded network of excellence Semantic Mining. Universal knowledge about entities of the world is represented within knowledge models (“ontologies of reality” i.e terminology systems), since known facts about concrete clinical cases (susceptible to error since they are assumed or suspected or be only true in today’s vision of things) are represented in information models (ontology of information). Whereas the ontology provides the framework of types/classes along which individual objects can be classified (e.g. pneumonia, fever) the information model determines the selection of what is described, its granularity, as well as its context (e.g. how the body temperature is measured, whether the diagnosis is speculative or definite). “Information models” describe the EHR content whereas the “ontologies of reality” describe real entities with taxonomies and (formal) descriptions. The domain content models (e.g. archetypes, CDA r2) can have mappings to any or all ontologies of reality. Extracts from the electronic Health record, based on common shared domain conent models are proposed as a means to exchange information between different heterogeneous health information environments.
c) Indexing structured data: binding clinical language units to terminology system Class labels of formal ontologies are not to be mistaken for actual terms. In order to map content of clinical language with classes of a formal ontology, it is necessary to provide a link between an ontology and a dictionary where each dictionary entry corresponds to a domain term and is linked to one (or to two or more, in the case of polysemous terms) class in the ontology. Synonyms are linked to the same node. Such a dictionary is a separate data structure and it is not an integral part of an ontology itself.
3) Terminology server To achieve interoperability within and across disparate healthcare IT systems, terminology resources for indexing both narrative and structured clinical information need to be synchronized across the various applications (at a local, or regional, or national, or international level).
For clinical structured information, terminology binding issues are addressed at the level of Value Sets defined in domain content models (content template based on CDA r2, archetypes, DICOM objects). These Value Sets need be synchronized across the various applications. The Integration Profile “Sharing Terminology Value Sets” provides only mechanisms for Value Sets synchronization across IT systems. In addition, the “Terminology Sharing” profile addresses the issues of 1) terminology services dedicated to support the terminology binding process while creating a new domain content model and 2) distributing and synchronizing updated domain content models.
For narrative clinical documentation, the “Terminology Sharing” profile will define terminology services supporting the indexation of narrative documents and enhancing search engines capabilities within narrative documents. The “Terminology Sharing” profile will provide solutions for a terminology server to distribute and update across IT systems a whole terminology resource, as complex as SNOMED CT for example (or subsets of a terminology resource).
The “Terminology Sharing” profile also addresses the issue of synchronization of terminology resources between the editor of the resources (e.g SNOMED CT IHT SDO) and terminology servers.
This requires that 1) terminology editors (Terminology Creator Sources) distribute terminology resources (Code Systems) to terminology servers (Terminology Sources), 2) terminology servers (Terminology Sources) maintain relationships between different terminology resources (Code Systems) and between Code Systems and Value Sets of different domain content models and also distribute updated inter-related Code Systems and Value Sets to IT applications (Terminology Repositories/Registries, Terminology Consumers)
Existing issues:
1) There is a wide variety of Terminology Resources (Code Systems) that have different level of complexity (from simple lists of terms to description logic-based ontologies)
2) Terminology services should be useful for applications providing indexation solutions of structured data but also for applications providing indexation solutions of narrative documents. Such an application may require other resources than terminology resources (lexicon, textual synonyms, etc). These are out of scope for this proposal.
Key Use Case
Use case 1.1: Importing a whole terminological resource (Code System). An entire Coding System is sent by the Terminology Source to subscribing systems (Terminology Registry/Repository). These systems must import the Coding System.
Use case 1.2: Importing update of terminological resource(Code System). If a subset of concepts/terms/codes of a Coding System is added, removed or changed, the full Code System is not sent to the Terminology Registry/Repository but only those parts which have changed. Concepts/terms/codes which have been removed from the Code System are not to be used by the receiving system any more; they should not be deleted but be flagged as disabled/invalid for backward compatibility reasons. New added codes may be used from the effective date/time given in the transaction.
Use case 1.3: Exporting a whole terminological resource (Code System). An entire Coding System is sent by the terminology server (Terminology Registry/Repository)to subscribing systems (Terminology Consumers). These systems must import the Coding System.
Use case 1.4: Exporting update of terminological resource(Code System). If a subset of concepts/terms/codes of a Coding System is added, removed or changed, the full Code System is not sent to the Terminology Consumer but only those parts which have changed. Concepts/terms/codes which have been removed from the Code System are not to be used by the receiving system any more; they should not be deleted but be flagged as disabled/invalid for backward compatibility reasons. New added codes may be used from the effective date/time given in the transaction.
Use case 2.1: Authoring a Value Set. While creating a new Value Set, if the Value Set Source needs a new unknown concept/term/code or subset of concepts/terms/codes the Value Set Source queries the Terminology Source (or the Terminology Registry/Repository)for the new code or the new subset.
Use case 2.2: Updating a terminology set. If a local application needs a new unknown concept/term/code or subset of concepts/terms/codes the Terminology Consumer queries the Terminology Repository/Registry for the new code or the new subset (while designing a questionnaire for structured data entry, while designing a text mining algorithm for narrative document indexation ?). If the concept/term/code or subset of concepts/terms/codes is not available in the Terminology Repository/Registry, the terminology server queries the Terminology Source for the new code or the new subset.
Standards & Systems
The CTS defines the minimum set of functions required for terminology interoperability within the scope of HL7’s messaging and vocabulary browsing requirements. The LexGrid model has been selected by HL7 as the vocabulary model in which the HL7 vocabulary will be represented. In the IHE Laboratory TF (LAB-TF), the exchange of code sets and associated rules shared by multiple actors is taken care of by a dedicated integration profile called “Laboratory Code Set Distribution” (LCSD) which is based on HL7 V2.X Master Files.
Technical Approach
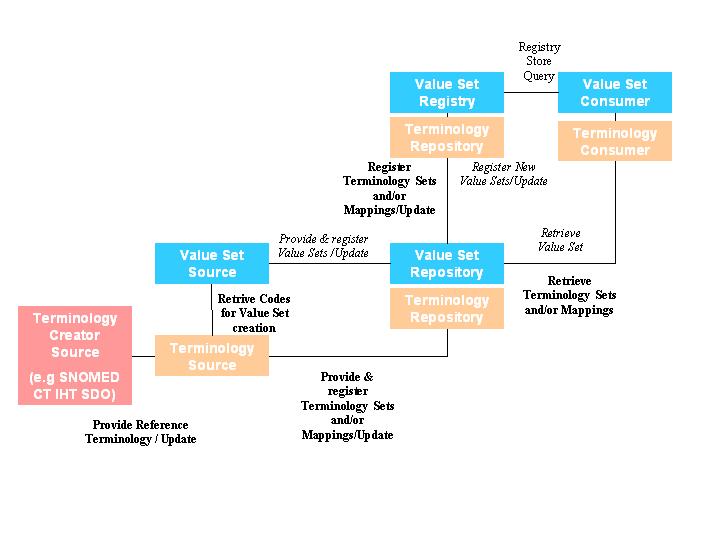
A similar approach as the ITI-XDS is adopted for the distribution of terminology resources.
Existing actors (actors in blue from integration profile "Terminology Value Sets Charing")
Value Set Source (defines relationships between Value Sets and Code Systems), distributes updated Value Sets to IT applications (Terminology Repositories/Registries) Value Set Repository
Value Set Registry
Value Set Consumer
New actors
Terminology Creator Source (maintains and distributes a terminology resource (Code System))
Terminology Source (maintains updated terminology resources, relationships between different terminology resources (Code Systems) and distributes Code Systems to IT applications (Terminology Repositories/Registries and Terminology Consumers).
Terminology Repository
Terminology Registry
Terminology Consumer
Existing transactions (transactions in italic from integration profile "Terminology Value Sets Sharing")
Provide & Register Value Sets
Registry Query Store Value Sets
Retrieve Value Sets
New transactions (standards used)
Provide & Register Terminology Sets and/or Mappings
Update Terminology Sets and/or Mappings
Registry Query Store Terminology Sets and/or Mappings
Retrieve Terminology Sets and/or Mappings
Provide & Register a Reference Terminology (from the editor)
Impact on existing integration profiles
<Indicate how existing profiles might need to be modified.>
New integration profiles needed
<Indicate what new profile(s) might need to be created.>
Breakdown of tasks that need to be accomplished
<A list of tasks would be helpful for the technical committee who will have to estimate the effort required to design, review and implement the profile.>
Support & Resources
In France many hospitals are deploying new generation of Clinical Information Systems and the government conducts a project of a national scale a PHR (Personal Health Record). The GIP-DMP (french public organization of the PHR), the GMSIH (The Association in Charge of the Modernization of the Healthcare Information Systems), researchers from the INSERM (The National Institute of Health and Medical Research), CNRS (National Center for Scientific Research), and the Association of Hospitals of Paris containing more then 40 hospitals are willing to put efforts into this profile. There is a strong interest from the industry side, namely companies such as Thales, Mondeca, and DBmotion.
Risks
<List technical or political risks that will need to be considered to successfully field the profile.>
Open Issues
The “Terminology Sharing” profile could provide solutions for a terminology server to distribute and update across IT systems: - Mapping between terminology resources (allowing for example classification and aggregation of data at different level). - Mapping between reference terminologies (“officially” distributed) and interface terminologies and/or local terminologies [17,18]. - Domain content models and corresponding Value Sets
The “Terminology Sharing” profile could provide solutions - To maintain the consistency between post-coordinated items - To synchronize Terminology/Value Sets/Mapping Registries/Repositories across Terminology Servers (e.g the local Terminology Server of an hospital and the national Terminology Server of the personal EHR)
References
[1] Kalra D. Electrnic Health Record Standards.Methods Inf Med 2006; 45 Suppl 1:107-13.
[2] Schadow G, Mead CN, Walker DM. The HL7 reference information model under scrutiny. Stud Health Technol INform 2006; 124:151-6.
[3]Cimino J.J. Terminology Tools: State of the Art and Practical Lessons. Meth Inform Med 2001:298-307.
[4]J. J. Cimino. Desiderata for controlled medical vocabularies in the twenty-first century. Methods of Information in Medicine, 37(4/5):394–403, 1998.
[5]R. Cornet R, N. F. De Keizer, A. Abu-Hanna. A Framework for Characterizing Terminological Systems. Methods of Information in Medicine, 2006; 45: 253-266.
[6]ISO/TC251 WG3. Standard specification for quality indicators for controlled health vocabularies; 2000 July. Report n° TS 17117.
[7]Rector AL. Clinical terminology: why is it so hard? Methods Inf Med. 1999 Dec;38(4-5):239-52.
[8]The OBO Foundry: http://obofoundry.org/, 2007.
[9]SNOMED Clinical Terms. 2007. International Health Terminology Standards Development Organization (IHTSDO) http://www.ihtsdo.org/.
[10]K. Spackman, G. Reynoso: Examining SNOMED from the perspective of formal ontological principles: Some preliminary analysis and observations. 72-80. In: U. Hahn (Ed.): KR-MED 2004, First International Workshop on Formal Biomedical Knowledge Representation, Proceedings of the KR 2004 Workshop on Formal Biomedical Knowledge Representation, Whistler, BC, Canada, 1 June 2004. CEUR Workshop Proceedings 102 CEUR-WS.org 2004
[11] Schulz S, Suntisrivaraporn B, Baader F. SNOMED CT's problem list: ontologists' and logicians' therapy suggestions. Medinfo. 2007;12(Pt 1):802-6.
[12]I. Horrocks, P. F. Patel-Schneider and F. van Harmelen, “From SHIQ and RDF to OWL: The making of a Web Ontology Language”, Journal of Web Semantics, 1(1), 7–26, 2003.
[13] Schulz S, Stenzhorn H.Ten theses on clinical ontologies. Stud Health Technol Inform. 2007;127:268-75.
[14]O: Bodenreider, B: Smith, and A: Burgun. The ontology-epistemology divide: A case study in medical terminology. In Achille C. Varzi and Laure Vieu, editors, Formal Ontology in Information Systems. Proceedings of the 3rd International Conference - FOIS 2004, pages 185–195. Amsterdam etc.: IOS Press, 2004 .
[15]A. Rector, R. Qamar, T. Marley. Binding Ontologies & Coding Systems to Electronic Health Records and Messages. In: Bodenreider O, editor. Formal Biomedical Knowledge Representation (KR-MED 2006) CEUR; 2006. p. 11-19.
[16]http://www.hl7.org/Special/Committees/terminfo/].
[17]C. Daniel-Le Bozec, O.Steichen, T. Dart, M-C Jaulent.The role of local terminologies in electronic health records. The HEGP experience. Medinfo 2007, vol 12, p.780-84
[18]Rosenbloom ST, Miller RA, Johnson KB. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc. 2006 May-Jun;13(3):277-88.