PaLM Conf Minutes 2023-June-13-14
Jump to navigation
Jump to search
Attendees
| Name | June 13 | June 14 AM | June 14 PM with DICOM |
|---|---|---|---|
| Gunter Haroske | X | X | X |
| Jason Lowder | X | X | X |
| Jim Harrison | X | X | X |
| Alessandro Sulis | X | X | X |
| Riki Merrick | X | X | X |
| Ruben Fernandes | X | X | X |
| Mary Kennedy | X | X | X |
| Raj Dash | X | X | X |
| Jans Hutrup | X | X | X |
| Megumi Kondo | X | X | X |
| Ralf Herzog | X | X | X |
| Marcial Garcia-Rojo | X | ||
| Tamas Sargar | X | ||
| Gabor Garanyi | X | ||
| Gabor Herman | X | ||
| John Jarvis | X | ||
| Markus Herrmann | X | ||
| Micke | X | ||
| Darren (ICX) | X | ||
| Sam Spencer | X | X | |
| Kevin Schap | X | X | |
| Bar Diricx | X | ||
| Dan Rutz | X | ||
| Ian Gabriel | X | ||
| Maria de la Iglesia | X | ||
| Ty Usrey | X | ||
| Mirelva Drost | X | ||
| Szabolcs Somoskeoy | X | ||
| Kenichi Tamama | X | X |
June 13, 2023
- Agenda review – no changes, except Juergen will not be here to talk about Gemini project
- Use cases - https://thecap-my.sharepoint.com/:p:/g/personal/salaver_cap_org/EWLwVv7MC-hCpi1HJciiV5wBNTDxZofgKcGYYxB06L_L-Q?e=HNcfhC
- SET discussion:
- CAT acceptance criteria
- Actors should be able to assert conformance for the use cases / scenarios that are related to the other IHE PaLM profiles their system supports – at minimum for the sender.
- For the receiver of SET - should it support all scenarios?
- If we ask the receiver to have logic to reconstruct the event metadata, then we may need to rethink if they should support all or just the ones that are related to the use cases their system supports.
- At the least it should be use case (more than one scenario) for 2 different vendors for both actors.
- Related profiles should be listed in the description of the profile for the CAT.
- For message structure should we define different MSH-21 identifiers to make it easier for Gazelle – similar to how we identify optional profiles in LAW?
- That might also help with vendors being able to declare the integration statement.
- CAT acceptance criteria
- What systems are supporting this profile?
- Could be any system that is interested in knowing more about the state of the specimen – LIS / Analyzer / Labeler / similar to an audit function in other use cases.
- European CAT
- DPIA Test Case update
- Alessandro is waiting for Anne-Gaelle to show him how to upload the DPIA test cases into Gazelle.
- Waiting to find out how many vendors are planning on testing for DPIA.
- Also looking for other systems for LTW testing to support the full profile.
- DPIA Test Case update
- Reviewing the test cases at high level – not in detail
- https://docs.google.com/spreadsheets/d/1cOIwUuHTwTnQbCSejUFyHQv_NlQQWQ5DBxnschNoJiw/edit#gid=1143561967
- Specimen container
- First test case may not be applicable to private labs, so should the pre-labeled step be optional?
- The difference is the perspective of the actor where the tracking is done (private lab sends out the labeled container, but may not be the collector (so they cannot create the message as the SEI) - for tracker that system MUST be able to support ALL aspects, else we cannot test all scenarios for the use case.
- Trackers may be at each organization; tracker should definitely support all.
- Monitor instructions
- First, inspect for correct message
- Ideally use the proxy to capture the message as part of the system-to-system check.
- Second, inspect for test case specific content.
- Example: the actual specimen and container ID declared by the labeler are what is used in both messages.
- First, inspect for correct message
- How do you make the sure the identifiers are unique?
- Ensure we have identifiers for each of the instances of the systems that assigned the ID.
- Ideally use OID, each org can get a root OID, so they can assign them to organization parts, system etc.
- They may not be publicly searchable, but at least the numbers should be unique, so when appended to the actual identifier, then the entire ID should be unique
- Ensure we have identifiers for each of the instances of the systems that assigned the ID.
- First test case may not be applicable to private labs, so should the pre-labeled step be optional?
- Testing conditions
- Always test for successful collection
- Specimen archive tracker only for the successful collection
- When the lab does not provide the containers
- Container prep + collection
- Must test all 3
- Always test for successful collection
- Gazelle provides message validation using profiles created in NIST IGAMT
- Gazelle also offers simulators
- Gazelle documentation / training
- Next steps:
- Identify the real-world scenarios within each of the use cases that we may need to support in testing and provide the name for each one.
- For official publication of the message structures, we are using will be in V2+, but V2+ the first version will be the same as the 2.9.1, so this will be in the next version after that
- SET discussion:
- DPIO - https://thecap-my.sharepoint.com/:p:/g/personal/salaver_cap_org/EQxFSq6O-YpIryjMsRMouIwBNKuHO8g5EZMS0DVNDDOD6A?e=piMERO
- DICOM questions
- What is the current data model for pathology compared to the radiology model?
- What data elements are needed in HL7 for meta-data?
- Make sure we explicitly push for the use of the assigning authority –
- If I have a GTIN+identifier – could use the GS1 OID.
- Mapping to the DICOM model
- Every new scan is a new series, assigned by the scanner.
- The scanner uses the study ID given from the order = case ID.
- Each next step is the Procedure Step
- Where to put the block?
- Part of the specimen preparation description?
- There is some imaging happening as part of block preparation to ensure that slides with the tissue do not go missing.
- Images from the same slide (rescan at different resolution)
- The scanner ALWAYS assigns UID for each new slide at a different point in time – you know they are the same based on the study ID.
- The barcode provides the linkage to the details that are in the DICOM header meta-data.
- DPIA supports sending instructions to the scanner on how the scan should be done as part of the order code.
- Specific scanning protocols – we need someone to create the standard choices – maybe put into LOINC or SNOMED CT observables?
- How to track which slide the pathologist looked at.
- What about storing of the heatmap longer term?
- Each heatmap is user specific – maybe this should be part of evidence creation?
- Which part of the slide was retrieved (and if it was in the viewer)
- Example: Ideally need to know I have looked at lymph nodes at 4x and everything else x2.
- Annotations on slides to identify the area of interest for second opinion - the DICOM supplement 222 covers that, but several elements seem to be missing (not being implemented because clients are not pushing on that).
- Also need to have the evidence creation profiles to standardize who to help with AI = DPEC.
- Report out the calculations done in PACS.
- Distance to margins – this is currently copied into the report – need to differentiate which system falls under IVD rules (regulation).
- Who has the worklist of the pathologist – separate actor?
- Case level is in LIS.
- Image level is in PACs.
- Viewer
- Propose we will use the terms “information creator” and “information receiver” so we can accommodate different systems that want to play these roles.
- Dynamically learn what information is available in systems that go to the network – may need to be very complicated.
- XSD in combination with the payload (CDA) with structured data – Sectra has done this in the islands.
- Could we also use a broadcast message instead of the query?
- Just send all – not sure if all systems could handle that and also that is not desired by the systems for privacy reasons – they will want to authenticate against each actor to each document type.
- The DP-AT white paper covers a lot of these scenarios.
- Should there be a document registry?
- DICOM Annotation
- Missing the type of analysis the annotation is related to.
- Have the image object.
- Have some SCT codes to represent some of the attributes the annotation represents.
- Need to map out concrete use cases to find out where all the required elements are.
- Biomarkers is a good use case, since multiple different things are measured.
- Now we are marking off the areas that are not of interest – negative heatmaps.
- Maybe create annotations that need to be presented to a pathologist since not clear to AI.
- What if annotations are within another annotation (special relationship) or partial overlap – cannot currently ask for overlap of those 2 annotations?
- Measurements currently are only for specified coordinates – no way to express measurement from a specific element of interest.
- Sectra has annotations for tracking lesions in image instances in radiology:
- No volume support in radiology.
- Based on special coordinates to the SOP (so should still work on WSI with different magnifications).
- Sectra has annotations for tracking lesions in image instances in radiology:
- How does this work for ejection fraction in ECHO – that would have to be done off comparison of different images – how are those annotated?
- Is that not in the evidence creation profile territory?
- Do we need to be able to “order” a scan function in relation to a particular annotation?
- Create 2 transactions in DPIO; one for the more complex information, then that might work.
- That would be a request in the LIS for creating a new slide and then request the new slide for the invasive focus where you want to count the cells.
- Missing the type of analysis the annotation is related to.
- Do we have annotations about clinical data related to focus regions (ROI) in images?
- DICOM already has elements for all of this – but all of those are optional, most vendors currently only pass the barcode as the only DICOM required element.
- If all the diagnostic information is in an image, then that can become a privacy issue.
- If the order starts in the PACS, then the order needs to go to the scanner and the LIS – or just send it to the acquisition manager?
- Quant analysis is done in AI – the ROI is marked by the pathologists (though in the future we can also expect the AI to identify which areas may be ROI). For re-cuts you cannot mark it in the order – so it is more of a reminder on where to go look – once you have the slide, then you can identify the ROI for the follow up AI order.
- Carry the ROI from the first image for co-registered images to the related area in the new slide.
- Reference the ROI DICOM annotation ID in other messages to the other actors, or rely on the image display actor to coordinated this when accessed?
- This is more of a workflow support action.
- If you want to convey:
- Suspicious for invasive tumor.
- Or just say this is ROI and have the order indicate what you want to have done.
- Add a comment to help the other pathologist to understand why something was ordered.
- Annotation needs to go into both DPIO and DPEC.
- Issues:
- Interactive workflow based on pathologist review.
- SOP driven workflows.
- Which system handles the workflows and who do I have to tell- LIS or PACS?
- LIS knows most of the workflow (for physical work for sure).
- PACS (or associated systems like AI) generate data values that must be sent back to the LIS.
- If we design a new workflow, we should have all the information in one system that is needed to render a diagnosis.
- We should think of annotations as observations that are either instructions or additional data.
- DICOM questions
- Can we model everything we talked about in LAB-1?
- Yes – should be able to add in the annotations as OBX under OBR.
- We can package all DICOM meta data elements to an OBX that are being sent under the OBR.
- It’s not elegant or efficient as this creates long messages, but it is easy to implement.
- Where do you put the annotation category and type from DICOM and reason for annotation into the OBX?
- Does this need a group of OBX segments for 1 annotation?
- Or, can we send one OBX with a pointer to the DICOM file and NTE with the annotation reason?
- Or, send one OBX with ED of the DICOM image (that would be overkill).
- Yes – should be able to add in the annotations as OBX under OBR.
- Next step:
- Take each use case and map each one into the LAB-1 message to test this approach.
- Proficiency Testing Profile (PTW) - https://thecap-my.sharepoint.com/:p:/g/personal/salaver_cap_org/EbLelaX_jAVDiOaX5s7H-i4B9CtsrdNgHJPLkYTUjdWAQQ?e=v48tsG
- Raj started writing the introduction.
- Hope to get the details worked out for next year on the messaging side.
- Will separate the additional questions around lab operations.
- Provide the result that are automatically reported AND have the final report in the portal.
- Potentially add LDA?
- eDOS?
- Similarily in IHE we have LCSD.
- Could present that in the manual in the web portal as the selection list for PT enrollment.
- Consider using the FHIR catalog for this, might need to extend some of the data elements.
- FHIR catalog does include instrument / CLIA setting etc.
- LIVD catalog includes the instrument and reagent and their combinations, but not currently the CLIA setting, though in the expanded excel version that is already tracked.
- Expanding LTW and LAW
- Evaluate how to improve messaging around instrument / reagent / method detail.
- Mapping to CAP PT elements
- LAW has instrument ID and type (OBX-18) and reagents (in INV segment).
- Some CAP specific elements would need to be evaluated further.
- Green has spot in v2.5.1 without pre-adoption.
- Grey – maybe have a spot.
- Kit ID (maybe able to get from the specimen ID).
- CLIA Lab number – map to OBX-23.10.
- LIS and version – map to SFT.
- More information is probably needed from the CAP.
- When would we have LIDR?
- Summarize what LIDR is.
- Explain about the LIDR elements that we need.
- How do LOI and LRI compare to LTW?
- A lot more detail around implementation.
- Harmonization indicator discussion.
- FHIR
- Discussed on O&O calls.
- Gemini project
- How to create results using FHIR?
- FHIR messaging.
- FHIRcast project.
- FHIR subscription to task.
- FHIR notifications.
- V2-FHIR mapping.
- This is waiting to go to ballot.
- German Medical Informatics Initiative
- Sharing med data between academic hospitals.
- Defined a core data set in medicine for routine and research.
- Person / Dx / lab / procedures / medications / encounters
- Pathology report was an extension.
- Used the IHE CDA based APSR as basis.
- Used IPS as reference standard.
- Published in April 2023.
- DiagnosticReport as the base and bundled it with composition to create a document.
- There is an observation-based model where group observations replace the sections in the CDA.
- That makes searching for individual observations problematic if folks do not complete all the remaining.
- Some information of the clinical information section is now in the ServiceRequest.supportingInfo.
- Links: https://simplifier.net/MedizininformatikInitiative-ModulPathologie/~introduction and https://simplifier.net/guide/medizininformatikinitiative-modulpathologie-implementationguide2?version=current
- For SNOMED CT concepts for pathology, we need to work with Scott Campbell’s group.
- Discussed specimen yesterday – FHIR specimen resource will need to improve over time.
- FHIR catalog is going to be published: https://build.fhir.org/ig/HL7/fhir-order-catalog/.
- Alessandro working on profiles for biobanking for a FHIR sample locator.
- Querying for samples of specific kind is available across biobanking
- IHE LAW and ILW and LTW need to be re-examined.
- Device Identifiers in IHE profiles:
- OBX-18 (using EI) currently supports instrument model / manufacturer / serial number.
- MSH-3 (using HD) so could use GTIN there as well.
- We would have to ask for NEW Universal ID types, so we can send the UDI in MSH-3.2 and populate MSH-3.3.
- Is INV-1 (using CWE) for the contributing substances? – currently pointing to HL70451, which only lists ‘ALL’.
- We could make GSI, HIBCC and ICCBBA codes in HL70451.
- UP-146
- The different organizations that assign device identifiers each has their OWN format of how they look, and you need to know which one is being sent so that you can properly parse the information contained in one.
- Seems you have to send either the full UDI or can send parts of the UDI in multiple elements, BUT not parts of the UDI, but not all concatenated a one element.
- Might want to check with Marti Velezis – invite her to one of the next IHE calls. (After meeting, Ralf reached out and we are aiming for August or September)
- Device Identifiers in IHE profiles:
- Agenda for tomorrow:
- Starting at 9 AM – going till 4:30 PM.
- Joint session with DICOM in the afternoon.
June 14, 2023 – PaLM only in morning
- Introductions and agenda review.
- Digital Pathology overview - https://thecap-my.sharepoint.com/:p:/g/personal/salaver_cap_org/EWXOENjkOhJEs-iMNQDz0BsBuwqHfuQ9dbmbpIHiHXFIWA?e=eRihDj
- DICOM header info and other info into V2 messages.
- Need to analyze.
- DPAT vs LAB-1.
- Broadcast vs support for query.
- Need to analyze.
- DPIA / DPIO
- Get more of the DICOM header information into DPIA.
- Add OBX segments to LAB-80.
- More likely barcode scanning and then request the info via LAB-81.
- DPIO needs to handle requests for extra slides / additional info = workflow etc., but for the DICOM header elements will be in DPIA for the image already ordered.
- DPIO should include production of the slide to the LIS.
- Once the slide has been created, then use DPIA to create the workorder steps for the scanner.
- DPOR is that still needed?
- Similar to LAB-3 in LTW.
- The issue here is that a regular lab does not have a PACS.
- We should create a glossary for all the PaLM transactions, with the list of use cases / transactions / actors as a look up https://www.ihe.net/resources/technical_frameworks/#PaLM :
 to explain which profiles are where in the TF – we could add that to the table).
to explain which profiles are where in the TF – we could add that to the table).
- Clinical order from EHR-s (LAB-1 / LAB-3) that is split into workorders (LAB-4/LAB-5), which are then split to workorder steps (LAW) = DPIA).
- Get more of the DICOM header information into DPIA.
- A PACS is a storage place that you can write info to (from scanner) and get info out (LIS request tumor margin annotation), but that is NOT an order.
- This would be the QDO to get the images / annotation and WADO to get them back – actor here is image viewer.
- We need to define the workflow for the pathologist.
- What needs to get looked at / can order new slides / look at derivative results off the images?
- We need integration between the image storage actor and the LIS; maybe use something similar to radiology workflow profile (separate transactions for RIS – PACS would be analog to LIS – PACS).
- We need support these situations:
- Ordering a NEW slide / physical asset / scan – needs to support cancelation and other status updates (PACS status are registered / in progress / completed).
- PACS creates an order (OML) and sends it to the LIS, who returns the placer order number for that new slide, once created (ORL) or LIS can send the order the normal way (DPIA).
- LAB-4 uses the OML; in this case all you need back is the ORL to identify that the new DPIA cycle is started.
- Outcome of this order eventually is a new slide or something that didn’t work out.
- Do we need LAB-5?
- This is sending the result stating that the initial slide is appropriate.
- Graphical viewer is what the pathologist is working in.
- We need a message to start the creation of the physical slide.
- Order filler (biospecimen manager) to acquisition manager starts the initial workflow.
- Image manager is where the pathologist is doing the work, so ideally from there should be able to request the recut of the slide, which is a result of looking at the specimen that then causes the re-run.
- Getting information pieces from already existing reports / images.
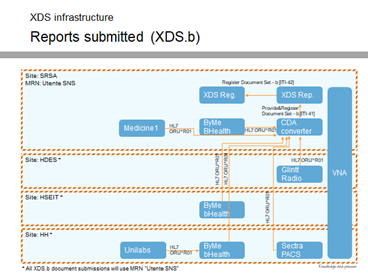
- If you want to have prior reports, consider using CDA, so that you can query based on the patient and is not something that is linked in the order. It would be sitting in the EHR-s, which in the US may be CCDs, but not all reports may be in the CDA format (but could propose to use XDS for this scenario); this is not currently an existing functionality.
- Requesting distance to margins that has already been measured (post evidence creation) from the annotated image – use QDO or FHIR query for discrete observation (both would be new to the LIS, so decide which would be better to work on – Ruben to report back)
- Example of this set up in Portugal:
- Ordering a NEW slide / physical asset / scan – needs to support cancelation and other status updates (PACS status are registered / in progress / completed).
- Sharing patient demographic data / updates (creation, merging) already handled in IHE PIR can use RAD-1, RAD-12.
- We need to communicate from the automation manager to the order filler suggesting a NEW order.
- Temporary identifier request from the PACS – may discuss using FHIR for this.
- How can an actor know what services an actor has?
- LCSD is for exchanging the tests that can be ordered from the lab (external).
- Subscription to specific types of messages trigger what system should get what message.
- LIS says block A is used up and cannot order on it; make it OOS; a work request can be rejected – NACK at application level in V2.
- OML^O21 (order centric) or OML^O33 (specimen centric) from automation manager to the order filler / similar for LAB-4 from filler to automation manager (LAB-50).
- How do we specify the stains on the order?
- SPM-6 is using an HL7 table with incorrect and insufficient values, so replace with DICOM value set using Stains / fixative / embedding medium from SCT in substance hierarchy.
- https://dicom/current/output/chtml/part16/sect_CID_8114.html
- https://dicom.nema.org/medical/dicom/current/output/chtml/part16/sect_CID_8115.html
- https://dicom.nema.org/medical/dicom/current/output/chtml/part16/sect_CID_8112.html - copyright issue?
- Site for browsing DICOM: https://dicom.innolitics.com/
- SPM-6 is using an HL7 table with incorrect and insufficient values, so replace with DICOM value set using Stains / fixative / embedding medium from SCT in substance hierarchy.
- How do we specify the stains on the order?
- DPIA CP:
- What do we need to add to the existing LAB-80 payload? We discussed adding IPC, which would mean updating LAB-80 to use v2.9 as the base structure or just pre-adoption the IPC segment.
- IPC supports the DICOM elements already.
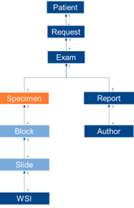
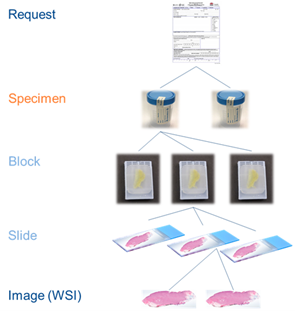
- Slide ID = SPM-2
- Block ID = SPM-3
- Container ID = SAC-3
- Accession ID = SAC-2 = SPM-30 = IPC-1
- We should doublecheck the mapping table Raj had against the DICOM mapping table we have in Gazelle.
- Specimen can be cut in parts (slices of breast material), from which the blocks are cut – if you leave out the part, then it must have the parts element as part of the block naming.
- SPM-4 SCT from specimen hierarchy.
- SPM-6 change values from DICOM value sets.
- Staining procedure for H&E, but not a single substance for H&E.
- What do we need to add to the existing LAB-80 payload? We discussed adding IPC, which would mean updating LAB-80 to use v2.9 as the base structure or just pre-adoption the IPC segment.
- Prep for DICOM
- Assigning authority in IDs.
- Consensus on pathology data model compared to Radiology model used in DICOM.
- Relationships between annotation – special only, or can you nest / base on landmark etc?
- Complex use case for data payload.
- Standard list for annotations – how to create in DICOM?
- Elements that are standard.
- Annotations for certain tissue types.
- How does DICOM document procedure steps like balancing the color profile to ensure scans from 2 scanners compare better?
- SCT clean up.
- Value set for stains / fixatives.
- Findings and Finding site.
June 14, 2023 – Joint PaLM / DICOM WG 26 Meeting (PM)
- Discussion topics:
- Discussions with DICOM WG:
- Consensus on pathology data model compared to Radiology model used in DICOM.
- Study = case.
- Series = WSI of a single slide.
- Different modality would be a different series.
- Procedure step is used in DICOM for workflow management.
- Request for image acquisition on a different modality – e.g. grossing vs slides.
- Can DICOM provide guidance on how to use procedure step?
- Radiologist worklist schedule.
- Used for billing.
- RIS is driving the workflow using procedure steps, so you can report status on each step.
- In pathology often need to look at the 3D gross specimen to better understand where the slice of the slide comes from so that this info can go back to the surgeon.
- Linkages via the specimen parent linking; DICOM has templates for that, but have not seen that at connectathons.
- Assigning authority in IDs.
- Every vendor requests a root OID and every DICOM image is traceable back to the instrument but not sure about the mechanism.
- Create a DICOM UID from UUID and append that to the 2.25 root, which guarantees that the ID is unique.
- DICOM supports different mechanism to define the assigning entity; these can be
- DICOMroot.<country id>.<vendor registration ID> and then append the ID.
- https://dicom.nema.org/medical/dicom/current/output/chtml/part05/sect_9.2.html .
- There are other places you can get a root OIDs.
- Identifiers are different – for those DICOM supports issuer sequence (optional).. https://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_10.14.html#table_10-17
- Can be different things, supports OIDs, namespace.
- PaLM profile can then further constrain the optionality.
- Relationships between annotation – is this special or can you nest / based on landmark etc.?
- Looking for standard list for annotations and how to create them in DICOM.
- Elements that are standard.
- Annotations for certain tissue types – see template discussion above.
- Annotate different cell types – label them using annotation.
- Can I then identify the nucleus of one of the specific cell types other than by spatial overlap of the annotations coordinates?
- Use annotation group for the label; can assign several to the group.
- But for each cell you can only have a single label.
- If the group label is using the right terminology, then can infer the right hierarchy for it, if using a hierarchical terminology, or you encode each step of the hierarchy you might want to use for queries.
- Markus has some examples for populating these annotation groups he can share.
- At the individual cell level can only support measurements – would have to come up with a schema to translate the numeric answers into qualitative evaluations.
- https://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.37.html#para_3917aaa9-40ef-443b-995c-c2df65159a39
- https://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.37.html#sect_C.37.1.2.1.2
- We may need a "Qualitative Evaluations Sequence" in addition to "Measurements Sequence" to assign multiple labels to individual cells.
- Other links:
- TCIA = https://www.cancerimagingarchive.net/
- https://dicom.nema.org/medical/dicom/current/output/chtml/part16/chapter_A.html#sect_TID_1500
- https://dicom.nema.org/medical/dicom/current/output/chtml/part16/chapter_A.html#sect_TID_1501 – this is a more general template for annotations – reasons for the choice is here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9712874/
- This template can be created for specific clinical use cases – might be good to map DICOM SR elements to FHIR mCode.
- Looking for DICOM connectathon at PATHVisions
- Get evidence creators involved.
- How do we deal with updates to annotations?
- New annotation is sent after the initial image has been deleted – what if that image was used in a report – can use audit trail described in IHE ATNA.
- Will need to figure out the audit trail and it it only works if you write the evaluation and reference the images (UID) you have actually used Is there a way to get to the ATNA artifacts as well.
- May be a FHIR use case, too.
- DICOM structured report has the status codes (prelim / final / verified / corrected).
- IHE Radiology created the AIR profile which deals with updates.
- SR does not currently shared back to the LIS.
- Difficult to get the structured data out – could create FHIR objects so they can be queried.
- New annotation is sent after the initial image has been deleted – what if that image was used in a report – can use audit trail described in IHE ATNA.
- Annotations should be locked – DICOM objects are supposed to be immutable, but not always implemented that way.
- Can create multiple files for each annotation group on an image.
- Imaging data commons should have many examples available.
- Markus created SR artifacts which include the diagnosis / there is a CP-2179 out to support separately encoding morphology and topography / finding site / finding = https://www.dclunie.com/dicom-status/status.html#:~:text=CP%202179,Pathology%20Annotation%20Codes
- Looking for standard list for annotations and how to create them in DICOM.
- Complex use case for data payload.
- Indicate ROI in image and drive workflow off that group of annotations.
- Need feedback.
- How does DICOM document procedure steps like balancing the color profile to ensure scans from 2 scanners are better comparable?
- SCT clean up.
- Value set for stains / fixatives.
- Should be all from the substance hierarchy – should this be modeled ss procedures instead.
- But for IHC and Immunofluorescence, it would be important to track the type of primary and secondary AB that has been used in relationship with the channel. This level of detail is in the lab, does it need to get into the DICOM object?
- There is a CP 2082 for dealing with this https://www.dclunie.com/dicom-status/status.html#:~:text=CP%202082,Add%20acquisition%20codes
- Consider how to address additional factors like concentration or time in fixative.
- Having a list of substances might still be helpful.
- May be helpful to categorize that the image is within the boundary of the intended use (FDA).
- Need to identify the right level of detail.
- Findings and Finding site.
- See mention above.
- Could we use CDISC to end codes as an alternate code system, since that is what FDA requires for submission for pharma?
- It would be good to get the FDA to use clinical standards.
- Value set for stains / fixatives.
- Consensus on pathology data model compared to Radiology model used in DICOM.
- Updates on DPIA
- Specify scanning parameters in tasks/workorders via the LIS or entered by the lab tech.
- Scanner includes bare minimum information in the barcode, then the image manager requests additional metadata from the LIS.
- Will share the link to the IHE Gazelle use cases for DPIA.
- Updates on DPIO
- Image manager should be able to:
- Query for information (this would be partly before the commit to the image manager (conceptually this would still be the acquisition modality actor, even if it in a different system – it is the functionality of having ALL data needed for commit to the image manager).
- Place a work order into the LIS.
- Image manager should be able to:
- Update from DPEC
- Looking for AI vendors to participate.
- AI vendors need to have metadata to know which elements they need.
- Need to let vendors know that being part of the standard development which they will need to standardize, if they want to work with more than one system.
- Hackathon would be a good idea – focus on something that has some clearance.
- Updates from DICOM
- Next Connectathon is planned to include AI and then want to transition the ownership of the Connectathon to the Digital Pathology Association. There are Connectathons in Europe, US and Japan and work on collaboration with IHE in the future.
- DICOM WG should focus on the creation of the standards.
- DICOM conformity issue still exists.
- After that we can focus on the annotations and AI improvement.
- Next Connectathon is planned to include AI and then want to transition the ownership of the Connectathon to the Digital Pathology Association. There are Connectathons in Europe, US and Japan and work on collaboration with IHE in the future.
- Discussions with DICOM WG:
- Next steps:
- Possible joint meeting at PATHVisions Oct 29, 2023 in Orlando, FL.